Note: those marked with an asterisk I am not 100% confident in my answer/reasoning.
Please feel free to discuss the problems and my proposed solutions below!
Quiz
*P1
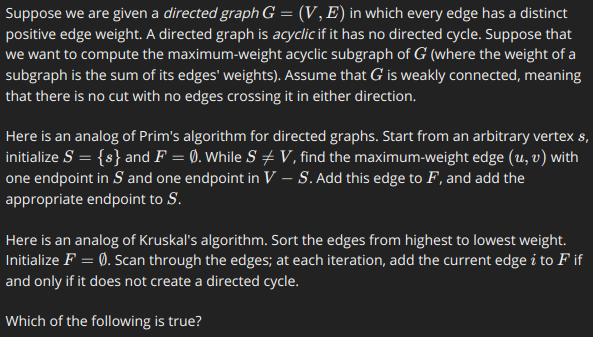
Both might fail to compute a maximum-weight acyclic subgraph because for any edge e that gets selected at any iteration, there could be a set of edges with smaller weights but sum to a value larger than the cost of e and still yield a DAG.
P2

Making the edge costs negative won’t prevent finding a minimum spanning tree in either algorithm, so both will work. However, since ties are broken arbitrarily, both algorithms may not produce the same minimum spanning tree.
P3

Kruskal’s algorithm proceeds by sorting the edges from least to greatest and then removes edges in order of increasing weight as long as they are not part of a cycle. The proposed algorithm is the inverse of Kruskal’s algorithm and will thereby produce the same partition of edges. Therefore this algorithm will always output a minimum spanning tree.
P4

For every edge in a MST, there is a cut for which that edge is the cheapest one crossing it. So any other spanning tree will have a maximum edge cost equal to or greater than that of a MST. Thus, an MST is always a minimum bottleneck spanning tree.
However, just because a tree has the smallest max edge cost possible does not guarantee that it is overall a MST. As a counterexample, consider a triangle with two unequal, relatively small edge costs and one really large edge cost. Two different bottleneck spanning trees can be made, but only one of them is a MST.
In other words, a MST is a subset, with unary cardinality, of bottleneck spanning trees of a given graph.
P5

All of them are correct. If an edge is in the MST and we decrease its cost, the MST won’t be affected. Likewise if we increase the cost of an edge outside of the MST.
If we decrease the cost of an edge, e, outside of the MST we can recompute the MST by examining each edge in the MST and see which one, if any, can be replaced with e. Likewise if we were to increase the cost of an edge inside of the MST, then we search the edges outside of the MST to see if an improved MST can be made.
Therefore, all possibilities listed can be done in O(m) time.
Programming Assignment
For both programming assignment problems, we need to make use of a unionFind datastructure. Here is my implementation below (maybe not the best implementation…):
C++
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
/******************************************************************************/
/******************************************************************************/
/* unionFind Header */
/******************************************************************************/
/******************************************************************************/
#include "../Graph/PrimNode.h"
#include <iterator>
#include <map>
using std::map;
#include <vector>
using std::vector;
using std::pair;
typedef int c_size_t; //size of cluster
typedef int leader_t; //value of leader node
class unionFind {
public:
unionFind(map<int, PrimNode*> nodeList);
/**
* Finds the head value of x's cluster.
* @param x
* @return Head of x's cluster, or -1 if x does not exist.
*/
int find(int x);
/**
* Unions the two clusters that i and j belong in, if they exist and are
* separate clusters.
* @param i
* @param j
* @return true if successful, false otherwise.
*/
bool unionClusters(int i, int j);
int numberOfClusters();
/**
* Returns a pair containing the ID of the cluster that i is in and the size
* of that cluster. This function calls this.find(int) and
* this.unionClusters(int,int) in turn calls this.sizeOfCluster(int), so
* it made sense to me to have sizeOfCluste also return the ID of the
* cluster so that unionClusters doesn't have to call find unnecessarily.
* @param i
* @return
*/
pair<leader_t,c_size_t> sizeOfCluster(int i);
private:
map<int, vector< PrimNode*> > clusters;
map<int, PrimNode*> nodeList;
};
/******************************************************************************/
/******************************************************************************/
/* unionFind Implementation */
/******************************************************************************/
/******************************************************************************/
unionFind::unionFind(map<int, PrimNode*> nodeList) {
// set nodeList field
this->nodeList = nodeList;
// populate clusters
vector<PrimNode*> cluster;
int id;
PrimNode* p;
for (auto& i : nodeList){
id = i.first;
p = i.second;
cluster = *(new vector<PrimNode*>);
cluster.push_back(p);
clusters[id] = cluster;
}
}
int unionFind::find(int x) {
try{
nodeList.at(x);
return nodeList[x]->getLeader()->getValue();
} catch(std::out_of_range){
return -1;
}
}
int unionFind::numberOfClusters() {
return clusters.size();
}
pair<leader_t,c_size_t> unionFind::sizeOfCluster(int i) {
pair<leader_t, c_size_t> result;
int n = find(i);
if (n != -1){
result.first = n;
result.second = clusters[ n ].size();
} else{
result.first = -1;
result.second = -1;
}
return result;
}
bool unionFind::unionClusters(int i, int j) {
pair<leader_t,c_size_t> iPair = sizeOfCluster(i);
pair<leader_t,c_size_t> jPair = sizeOfCluster(j);
//determine bigger cluster
int iClusterSize = iPair.second;
int jClusterSize = jPair.second;
// only union if both exist
if (iClusterSize == -1 || jClusterSize == -1)
return false;
int largerClusterID;
int smallerClusterID;
if (iClusterSize > jClusterSize){
largerClusterID = iPair.first;
smallerClusterID = jPair.first;
} else if(iClusterSize < jClusterSize){
largerClusterID = jPair.first;
smallerClusterID = iPair.first;
} else { //they're equal in size
largerClusterID = iPair.first;
smallerClusterID = jPair.first;
}
// if they have same leader, they are in the same cluster already, so skip
if (largerClusterID == smallerClusterID)
return false;
//set leader pointers of smaller cluster to that of the larger
PrimNode* newLeader = clusters[largerClusterID][0]->getLeader();
for (auto& i : clusters[smallerClusterID]){
i->setLeader(newLeader);
}
//copy stuff from smaller cluster to larger cluster
auto bitr = clusters [ smallerClusterID ].begin();
auto eitr = clusters [ smallerClusterID ].end();
auto here = clusters [ largerClusterID ].end();
clusters [ largerClusterID ].insert(here, bitr, eitr);
//delete smaller cluster
clusters.erase(smallerClusterID);
return true;
}
Parts 1
We’re asked implement the clustering algorithm and compute the k number of clusters needed for a spacing of 4. Below is my implementation, added to my undirected weighted graph datastructure from previous weeks. I also use my pqueue data structure to help sort and determine minimum edge costs. Just needed to feed a more appropriate comparator to the pqueue, which is also listed below:
C++
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
int UndirectedWeightedGraph::kspacing(int k) {
unionFind uf(nodeList);
pqueue<PrimEdge*, KruskalEdgeComparator<PrimEdge>> kEdgeList;
//copy over edges to new pqueue with appropriate comparator for the algo.
kEdgeList.insert(kEdgeList.begin(), edgeList.begin(), edgeList.end());
int cluster1;
int cluster2;
while (uf.numberOfClusters() != k){
cluster1 = uf.find(kEdgeList.top()->getFirst()
->getLeader()
->getValue());
cluster2 = uf.find(kEdgeList.top()->getSecond()
->getLeader()
->getValue());
// try to union the two clusters, if it works then we need to re order
// the pqueue.
if (uf.unionClusters(cluster1, cluster2)){
//reorders pqueue so that min edge between two clusters is on top.
kEdgeList.update();
}
}
return kEdgeList.top()->getWeight();
}
C++
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
template <class T> struct KruskalEdgeComparator {
bool operator() ( T* x, T* y) {
if (strcmp ( typeid(T).name(), "PrimEdge")){
if (x->differentLeaderNodes() && y->sameLeaderNodes())
return true;
if (x->sameLeaderNodes() && y->differentLeaderNodes())
return false;
return *x < *y;
}
else
throw std::string("KruskalEdgeComparator may only be used with PrimEdge objects!");
}
};
Runs in about 1.5 minutes. The sorting on almost every iteration is a point of potential improvement.
Part 2
In this part we’re asked to determine the largest value of k for a k-clusterring with spacing at least 3 with an input file containing a large number of nodes as binary values. The edges between nodes is implied by the number of differences between bits between each node. As such, computing every edge is computationally difficult, so a more clever implementation is required. For this, I decided to try the dictionary method where you go through each node, and determine all possible neighbors with edge costs 0, 1, and 2 while clustering together these nodes if they were in separate clusters. Below is my implementation:
C++
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
int UndirectedWeightedGraph::findKWithSpacingOfThree() {
unionFind uf(nodeList);
int n;
int neighbor;
cout << "Done with 0 buddies" << endl;
//iterate through each cluster looking for distance of 1 buddies
for (auto i = nodeList.begin(); i != nodeList.end(); i++){
n = i->first;
bitset<24> bn(n);
for (int d = 0; d < 24 ; d++){
bn.flip(d);
neighbor = (int)bn.to_ulong();
uf.unionClusters(n, neighbor);
bn.flip(d);
}
}
cout << "Done with 1 buddies" << endl;
//iterate through each cluster looking for distance of 2 buddies
for (auto i = nodeList.begin(); i != nodeList.end(); i++){
n = i->first;
bitset<24> bn(n);
for (int d = 0; d < 24 ; d++){
bn.flip(d);
for (int dd = d+1; dd < 24; dd++){
bn.flip(dd);
neighbor = (int)bn.to_ulong();
uf.unionClusters(n, neighbor);
bn.flip(dd);
}
bn.flip(d);
}
}
cout << "Done with 2 buddies" << endl;
return uf.numberOfClusters();
}
Runs in about 2.5 minutes.